![[OK]](tick.png) Basic Statistics
Basic Statistics
| Measure | Value |
|---|---|
| Filename | Chip-seq_Input_DNA |
| File type | Conventional base calls |
| Encoding | Illumina 1.5 |
| Total Sequences | 161531951 |
| Filtered Sequences | 0 |
| Sequence length | 36 |
| %GC | 40 |
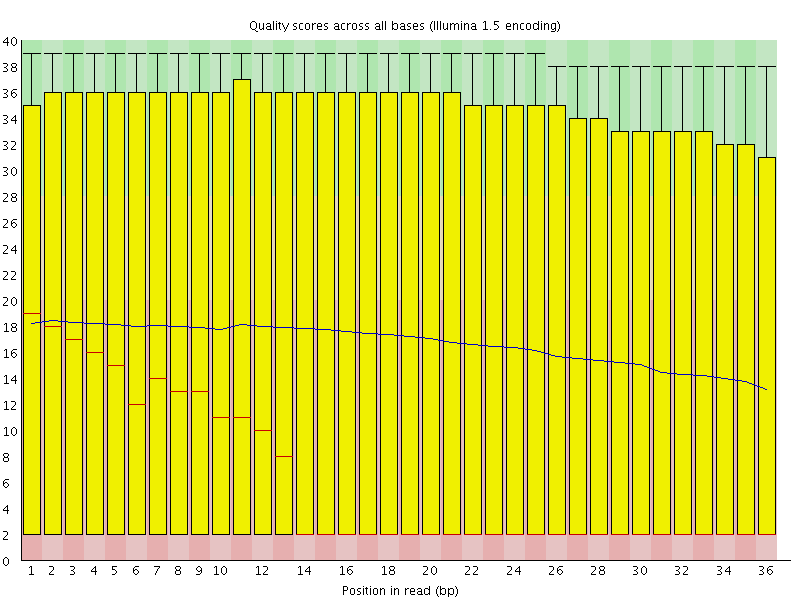
![[FAIL]](error.png) Per base sequence quality
Per base sequence quality
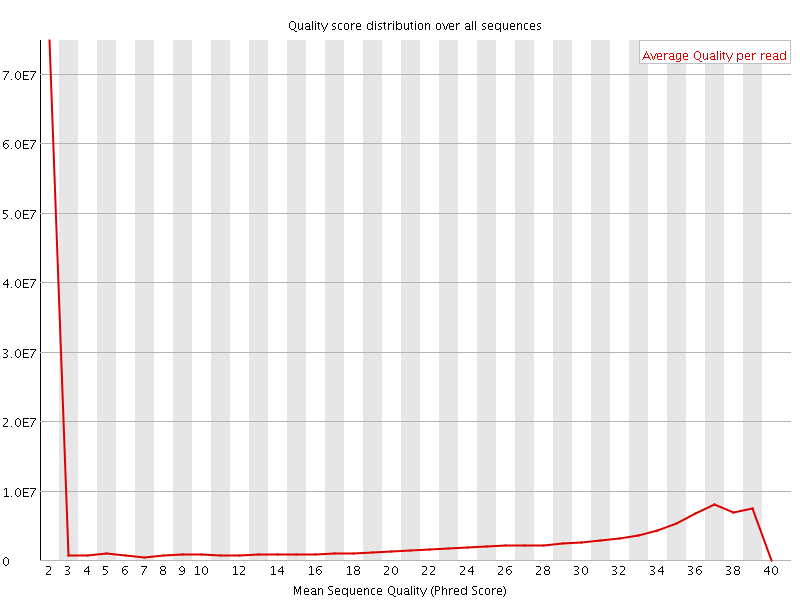
![[FAIL]](error.png) Per sequence quality scores
Per sequence quality scores
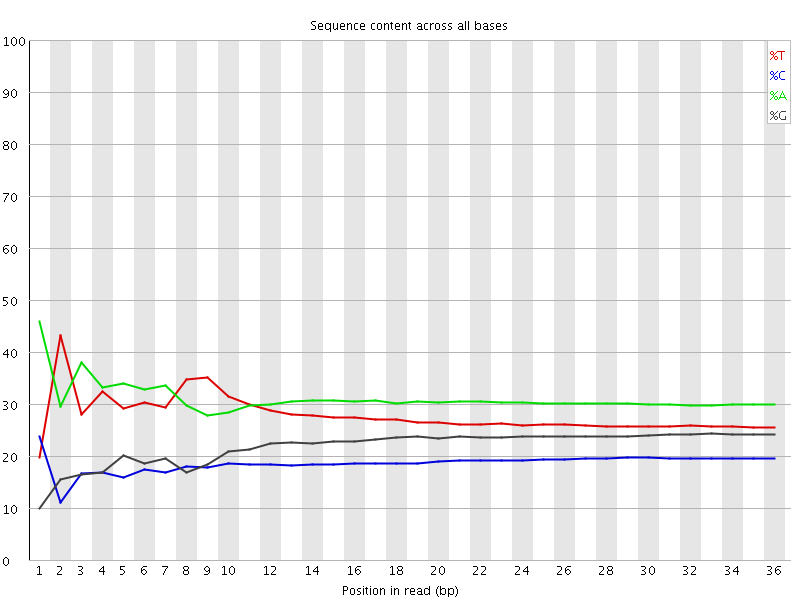
![[FAIL]](error.png) Per base sequence content
Per base sequence content
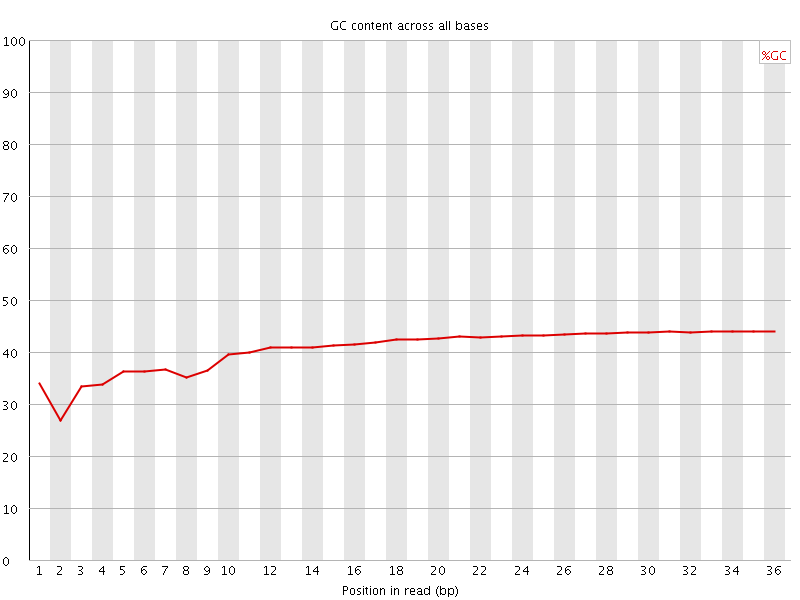
![[FAIL]](error.png) Per base GC content
Per base GC content
![[OK]](tick.png) Per sequence GC content
Per sequence GC content
![[WARN]](warning.png) Per base N content
Per base N content
![[OK]](tick.png) Sequence Length Distribution
Sequence Length Distribution
![[WARN]](warning.png) Sequence Duplication Levels
Sequence Duplication Levels
![[OK]](tick.png) Overrepresented sequences
Overrepresented sequences
No overrepresented sequences
![[FAIL]](error.png) Kmer Content
Kmer Content
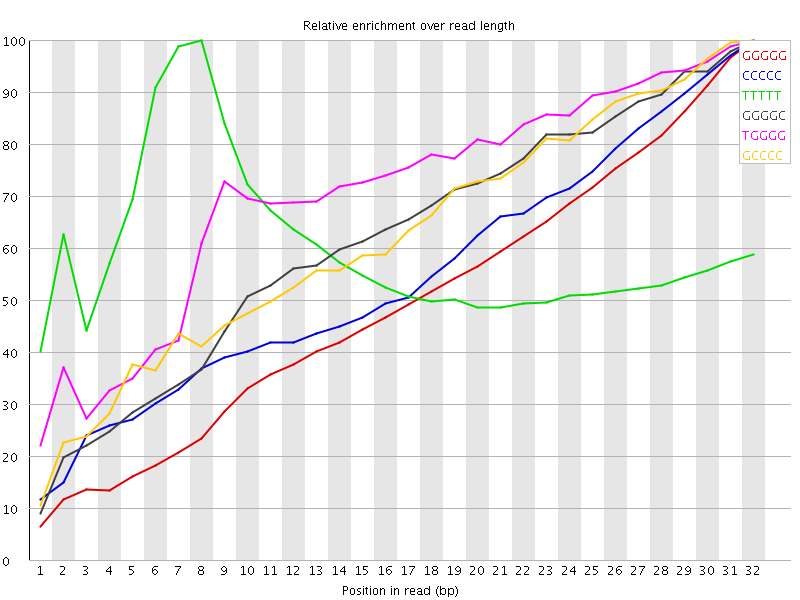
| Sequence | Count | Obs/Exp Overall | Obs/Exp Max | Max Obs/Exp Position |
|---|---|---|---|---|
| GGGGG | 17540390 | 6.708389 | 13.514825 | 32 |
| CCCCC | 7347310 | 6.185458 | 11.234757 | 32 |
| TTTTT | 29390610 | 3.3054726 | 5.5298185 | 8 |
| GGGGC | 7251345 | 3.2473602 | 5.238316 | 32 |
| TGGGG | 10725810 | 3.211429 | 4.5194926 | 32 |
| GCCCC | 4439560 | 3.1919096 | 5.0972133 | 32 |
| CTGGG | 8774125 | 3.0761318 | 3.794928 | 32 |